Female Hormone Tests: A Complete Guide to Assess Your Health and Fertility
Female hormones play a central role in virtually every stage…
Continue reading


Prenatal care is extremely important for maintaining the health of the pregnant woman and the baby. Relatively recent, the non-invasive prenatal screening test (NIPT) has helped transform obstetric practice, bringing even more safety.
In general, the main objective of prenatal care is to identify and treat diseases early that may be harmful to the well-being of the mother and/or the child.
Conventional first-trimester screening (ultrasound and serum biomarkers) allows the detection of the most frequent chromosomal abnormalities in the fetus during pregnancy. It is mainly aimed at detecting major aneuploidies (changes in the number of chromosomes), such as:
This screening has a sensitivity between 85-90% for these syndromes, with a false positive rate of 5% (1).
NIPT is the latest development in prenatal screening and is increasingly being offered in clinical settings to detect not only major fetal trisomies but also to provide a comprehensive analysis of the entire fetal genome, aiming to detect other aneuploidies and structural chromosomal abnormalities.
The test is based on methods such as next-generation sequencing (NGS) or other high-throughput analysis tools for free fetal placental DNA in maternal blood serum (2).
Keep reading to learn more about it!
NIPT is a non-invasive prenatal test used to screen for major chromosomal abnormalities in the fetus.
Technological advances in DNA analysis have made it possible to develop this test, which is based on the study of free fetal DNA present in maternal blood. It can study different chromosomal conditions with greater sensitivity and specificity without posing a risk to the mother and the baby.
Available NIPTs generally screen for the following chromosomal aneuploidies (3):
Some panels can also screen for aneuploidies in all other chromosomes.
Clinically significant fetal chromosomal abnormalities usually involve gains or losses of genetic material. These can vary in size from small chromosome segments (called “microduplications” or “microdeletions”) to entire chromosomes (i.e., aneuploidy) (4).
Whole chromosomal imbalances, as well as copy number variations (CNVs), can also be detected through the analysis of cell-free fetal DNA (cfDNA), including whole genome CNVs and targeted microdeletion syndromes (5) such as:
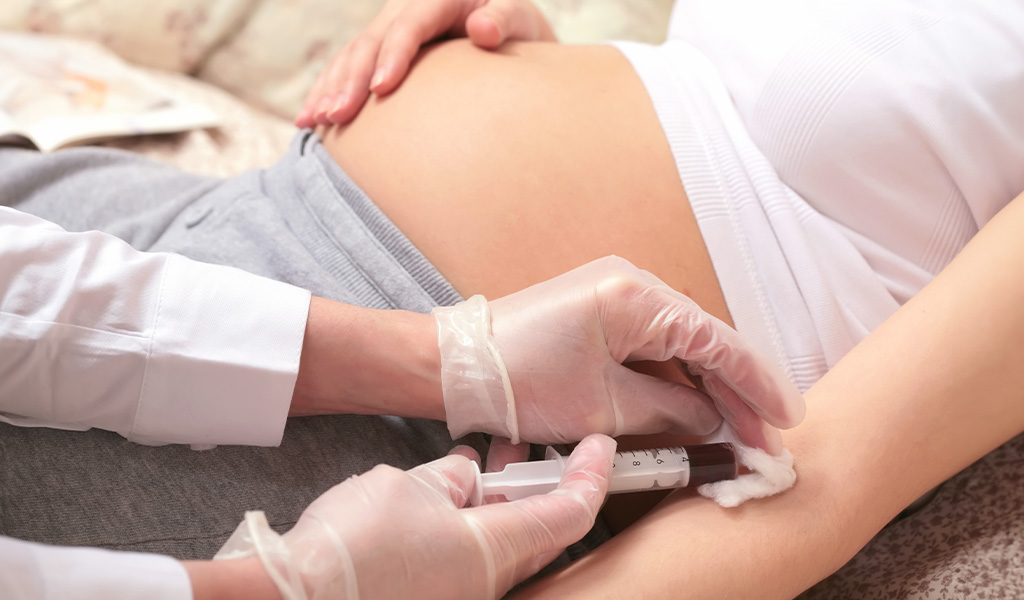
The NIPT test is performed by analyzing cell-free fetal DNA (cfDNA) in the maternal circulation, meaning fetal cells present in the mother’s blood. Thus, it is done through the collection of peripheral blood.
cfDNA (cell-free DNA) refers to DNA that exists as small fragments (less than 200 bp) in plasma or other body fluids, distinct from DNA contained in the nucleus of an intact cell, and released from all organs during a series of cellular processes (6). Maternal plasma cfDNA contains both maternal and fetal sources of cfDNA. The source of fetal DNA is the trophoblast (7), while the predominant source of maternal DNA is the hematopoietic system (8).
NIPT for fetal aneuploidy uses high-throughput next-generation sequencing (NGS) methods to quantify the proportional representation of each chromosome in the plasma cfDNA (9).
The proportional representation of each chromosome in plasma cfDNA reflects the chromosome size and the individual’s karyotype. In a euploid (46,XX) pregnant woman, a deviation from the expected chromosomal profile in plasma cfDNA due to excess or deficient cfDNA fragments of a specific chromosome suggests the presence of fetal trisomy or monosomy, respectively.
NIPT is a prenatal screening for chromosomal abnormalities, so it does not exclude the possibility of other genetic diseases (such as monogenic diseases caused by changes in a single gene).
Since NIPT is a screening test, the result should be evaluated by the requesting physician, within the clinical context, and with other laboratory or imaging findings of the patient.
High-risk results may reflect the presence of mosaicisms. Thus, the result can be confirmed by invasive tests as requested by the responsible physician and the patient’s desire.
A statistically significant variation in the count of cfDNA fragments for a given chromosome, commonly defined as a z-score >3, constitutes a high-risk result (4).
The fetal fraction (FF) is the percentage of total cfDNA in maternal plasma that is of fetoplacental origin. It is an assessment of both maternal and fetal cfDNA levels in maternal plasma.
Thus, the fetal fraction is a function of biological factors (i.e., maternal and fetal cfDNA levels in maternal plasma) and bioinformatics algorithms used to interpret DNA sequencing results, being essential in the quality control of non-invasive prenatal test (NIPT) results (4).
It is important to note that, although FF should be routinely calculated, there is still no consensus on whether it must necessarily be reported in the report.
Factors that may influence the fetal fraction include:
NIPT has been considered an advance in prenatal care for screening chromosomal abnormalities due to its clinical safety and ease of use.
Prenatal screening for fetal chromosomal abnormalities is performed to identify women at higher risk of having an affected fetus. It also allows for informed decisions about whether to proceed to diagnostic testing.
NIPT is indicated for pregnant women with at least 10 weeks of gestation (10 weeks + 0 days), in the following situations:
It can be performed in singleton or twin pregnancies, pregnancies resulting from in vitro fertilization (IVF), pregnancies resulting from IVF with gamete donation, and in cases of resorbed twins.
NIPT is a screening test, while a diagnostic test requires an invasive procedure, which can be performed between 11 and 14 weeks of gestation (10) by chorionic villus sampling (CVS) of placental tissue.
Alternatively, after the 15th week of gestation, the sample can be obtained by amniocentesis. Both procedures carry a small risk of causing miscarriage. The risk level is commonly reported as 0.5-1%, although recent studies suggest that the true procedure-related risk may be much lower (11).
Another issue is the rates of false positives and false negatives:
The false positive rate of NIPT for Down syndrome, for example, is generally 0.1%, meaning that the cfDNA test is positive for such a change, but the fetus is later determined to be unaffected. In a pooled study, the cumulative false positive rate was less than 0.4% (12).
Most false positives result from the presence of increased chromosome 21-specific DNA, the origin of which does not reflect the chromosomal composition of the fetus in the ongoing pregnancy.
Possible origins of this increased DNA include confined placental mosaicism (the presence of two or more karyotypically different cell lines present in the placenta and absent in the fetus), a resorbed twin, maternal mosaicism, and other maternal medical conditions (such as bone marrow or tissue transplant).
Although it is difficult to determine the true rate of false-negative results, these results are more common when the fetal fraction is low and/or when placental mosaicism is presente (13).
Overall, there are four main reasons for low fetal fraction (14):
The false-negative rates for the most commonly targeted aneuploidies are not sufficient to guarantee “diagnostic test” status. In this case, the diagnostic test used is fetal karyotyping.
Fetal karyotyping can be performed from chorionic villus sampling (CVS) or amniocentesis. Such collection procedures for karyotyping are invasive and pose a small risk to the pregnancy, with amniocentesis still considered the gold standard for invasive prenatal testing.
Conditions that contraindicate the use of the test are generally cases of fetal malformations, screening for less common aneuploidies, nuchal translucency equal to or greater than 3.5 mm, suspicion of fetal triploidy, and congenital infections, due to the very high risk of genetic alteration, in which direct indication for invasive fetal karyotyping is justified.
Discordant results between NIPT and invasive fetal karyotyping can occur due to biological processes, such as confined placental aneuploidy, a resorbed twin, maternal aneuploidy, or maternal câncer (14).
It is worth noting that this screening is a major advance in prenatal aneuploidy screening, but it is a screening test, not a diagnostic test, representing a risk for fetal chromosomal abnormalities, and confirmation should be done by chorionic villus sampling or amniocentesis. Additionally, genetic screening is optional and at the discretion of each individual patient, in conjunction with their doctor.
It is important to remember that NIPT interpretation should always be performed by the requesting physician based on the patient’s clinical and family history, together with other laboratory findings.
The non-invasive prenatal screening tests offered by SYNLAB are performed using next-generation sequencing Illumina®, paired-end (two reads are produced for each DNA fragment sequenced) of the entire genome through Whole Genome Sequencing (WGS) technology, allowing the measurement of free fetal DNA (cfDNA).
Since fetal DNA is smaller than maternal cfDNA, chromosome counts in smaller DNA fragments improve sensitivity and specificity even in cases of low fetal fraction, with an overall detection rate of 99.1% (95% CI: 95-99.9%).
SYNLAB offers the following NIPTs:
The neoBona test by SYNLAB detects:
Through WGS Paired-end technology + fetal fraction percentage. Suitable for singleton and twin pregnancies.
The neoBona Genomewide test detects:
Through WGS Paired-end technology + fetal fraction percentage. Suitable for singleton and twin pregnancies.
The test requires only a small sample of maternal peripheral blood, collected in a specific tube using a kit provided by SYNLAB, from the 10th week of pregnancy (10 weeks + 0 days). Before this, the fetal fraction is very low and there is a higher chance of test failure; however, there is no upper limit for gestational age (FF increases with gestational age).
The test involves analyzing fragments of free fetal DNA by sequencing, followed by quantification of DNA fractions of the examined chromosomes against a control standard, using a bioinformatics algorithm to release the result.
Results consistent with aneuploidies based on cfDNA are screening results and must always be confirmed by a diagnostic technique, such as fetal karyotyping or fetal DNA analysis (amniocentesis or chorionic villus sampling), before any other medical intervention. In these cases, it is recommended that the patient receives appropriate genetic counseling.
neoBona® is the first cfDNA paired-end based screening test that uses an innovative informatics algorithm and provides double verification of chromosome count data, generating the T-SCORE (trisomy score calculation) which integrates several parameters to provide reliable results even in cases of very low fetal fractions. This allows results to be obtained in the vast majority of cases (recollection rate is about 1.5%).
The T-SCORE takes into account chromosome counts, fetal fraction, fragment size distribution, and sequencing depth, thus quantifying the probability of fetal trisomy.
Moreover, neoBona’s paired-end WGS sequencing technology allows for a deeper and more comprehensive analysis of cfDNA compared to conventional single-end WGS technology, generating more efficient sequence counts, increasing the precision of the analysis.
If any risk for the assessed syndromes is detected, a free confirmatory test is offered to the patient, whose analysis is performed by sending material obtained through an invasive procedure (amniocentesis or cordocentesis).
The sensitivity and specificity rates for neoBona are as follows (1):
neoBona’s maximum specificity reduces the number of false positives to practically zero (<0.1%), avoiding a high number of unnecessary invasive procedures, while its sensitivity is above 99%, meaning that in practice, it is very similar to a diagnostic test, detecting 100% of occurrences.
In conventional screening, sensitivity reaches 90%, while specificity is 95% (with a 5% false positive rate), where out of every 100 healthy fetuses, 5 are incorrectly classified as positive and subjected to invasive procedures that endanger the mother and the fetus.
Accurate and up-to-date tests are essential for making more accurate diagnoses and better guiding treatments. SYNLAB is here to help.
We offer diagnostic solutions with rigorous quality control for the companies, patients, and doctors we serve. We have been in Brazil for over 10 years, operating in 36 countries across three continents, and are leaders in service provision in Europe.
Contact the SYNLAB team and learn about the available tests.
1) Cirigliano V, Ordoñez E, Rueda L, Syngelaki A, Nicolaides KH. Performance of the neoBona test: a new paired-end massively parallel shotgun sequencing approach for cell-free DNA-based aneuploidy screening. Ultrasound Obstet Gynecol. 2017 Apr;49(4):460-464.
2) Liehr T; Lauten A, Schneider U; Schleussner E, Weise A. Noninvasive Prenatal Testing – when is it advantageous to apply. Biomedicine Hub. 2017;2,(1), p1-11.
3) Badeau M, Lindsay C, Blais J, Nshimyumukiza L, Takwoingi Y, Langlois S, et al. Genomics-based non-invasive prenatal testing for detection of fetal chromosomal aneuploidy in pregnant women. Cochrane Database Syst Rev. 2017,Nov 10;11(11):CD011767.
4) Hui L, Bianchi DW. Fetal fraction and noninvasive prenatal testing: What clinicians need to know. Prenat Diagn. 2020,Jan;40(2):155-163.
5) Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 2015 Mar;212(3):332.e1-9.
6) Hui L, Maron JL, Gahan PB. Other Body Fluids as Non-invasive Sources of Cell-Free DNA/RNA. Advances In Predictive, Preventive And Personalised Medicine. Springer Netherlands. 2014. p.295-323.
7) Alberry M, Maddocks D, Jones M, Hadi MA, Abdel-Fattah S, Avent N, Soothill P W. Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast. Prenatal Diagnosis. 2007;v27,n(5),p.415-418.
8) Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016; v.164, n(1-2),p. 57-68.
9) Chitty LS, Lo YM. Noninvasive Prenatal Screening for Genetic Diseases Using Massively Parallel Sequencing of Maternal Plasma DNA. Cold Spring Harb Perspect Med. 2015 Jul 17;5(9):a023085.
10) Carlson LM, Vora NL. Prenatal Diagnosis: Screening and Diagnostic Tools. Obstet Gynecol Clin North Am. 2017 Jun;44(2):245-256.
11) Akolekar R, Beta J, Picciarelli G, Ogilvie C, D’Antonio F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015 Jan;45(1):16-26.
12) Practice Bulletin No. 163: Screening for Fetal Aneuploidy. Obstet Gynecol. 2016 May;127(5):e123-e137. doi: 10.1097/AOG.0000000000001406.
13) Zhang H, Gao Y, Jiang F, Fu M, et al. Non-invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound Obstet Gynecol. 2015,May;45(5):530-8.
14) Gray KJ, Wilkins-Haug LE. Have we done our last amniocentesis? Updates on cell-free DNA for Down syndrome screening. Pediatr Radiol. 2018 Apr;48(4):461-470.
15) Rava RP, Srinivasan A, Sehnert AJ, Bianchi DW. Circulating fetal cell-free DNA fractions differ in autosomal aneuploidies and monosomy X. Clin Chem. 2014 Jan;60(1):243-50.
Female hormones play a central role in virtually every stage…
Continue reading
The moment a woman discovers she’s pregnant, a mother is…
Continue reading
Preeclampsia is one of the most significant hypertensive disorders of…
Continue reading